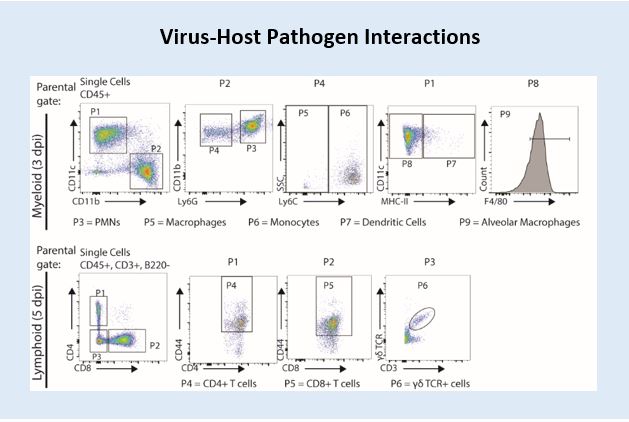
Virus-host pathogen interactions
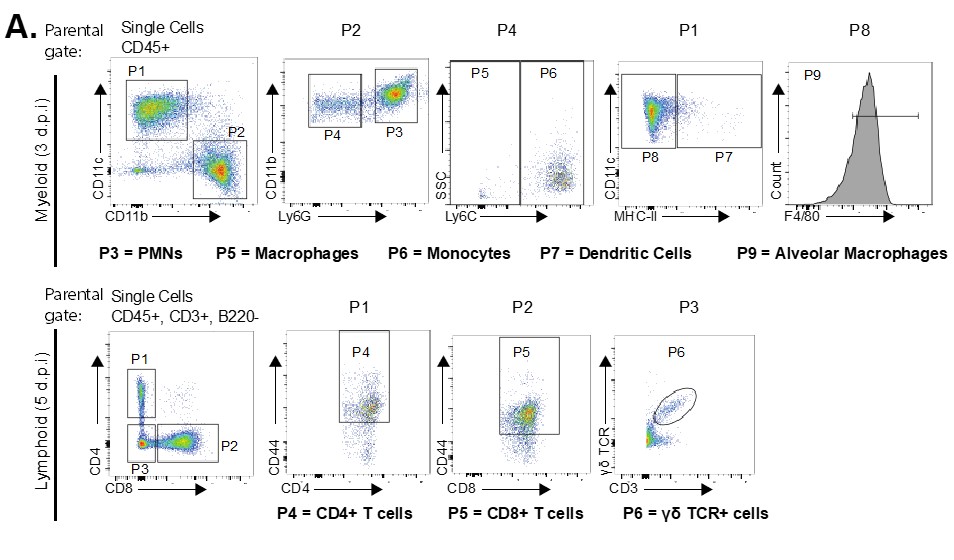
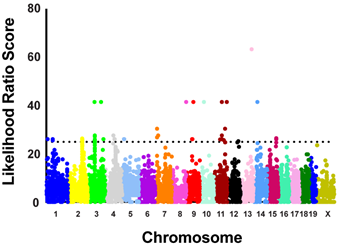
Viruses such as influenza virus can cause severe and fatal disease. Precisely how these events unfold during an infection are not fully understood. We use a combination of forward and reverse genetics to study virus-host interactions in vitro and in vivo. The forward genetic studies make use of the genetic variation between mouse strains and the impact of this variation on disease susceptibility and severity. We discovered almost 10 years ago that the DBA/2 mouse strains is extremely susceptible to the highly pathogenic avian H5N1 influenza virus. Genome-wide association studies identified several host factors that predisposed to severe disease including the complement factor 5 and a novel interferon induced protein 35 (IFI35). Next, we developed an IFI35 knockout mouse and found that IFI35 exacerbates disease after influenza virus infection. Detailed mechanistic studied found that the production of the host cytokine IL12p80 was reduced in the IFI35-knockout animals and that neutralization of this cytokine was beneficial for the mice. Future efforts will reveal how IFI35 regulates the expression or production of the key cytokine. Specific inhibitors of IFI35 or IL12p80 could be developed to treat severe influenza infections.
More recently, we discovered that the IFI35-knockout mice are also more resistant to Vesicular Stomatitis Virus (VSV) and we are now in the process of understanding how IFI35 promotes VSV replication and disease in vivo.
Additional projects in this program are
- Generation and characterization of novel gene-knockout mice. The genes in this project are in the same pathway as IFI35 or novel candidate genes from our forward genetic screens.
- Testing the effects of genetic variation in key host genes on disease pathogenesis.
- Quantifying the IL12p80 response in humans infected with influenza virus.
- Investigate the association between IL12p80 and severe disease in people.
Influenza virus vaccines
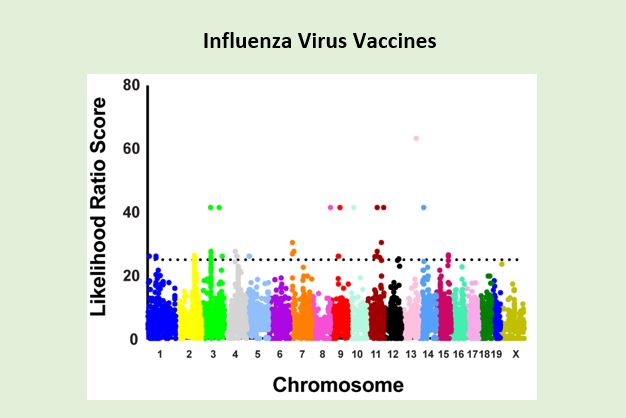
Vaccination is the key to an efficient health care system. It is estimated that over a million cases of childhood diseases have been prevented in the US over the past 100 years. Influenza vaccination is no exception and the CDC has estimated that over 40,000 lives have been saved between 2005 and 2014. However, the ability of the influenza virus vaccine to protect against influenza virus infection is suboptimal. The research in my laboratory is aimed to develop novel vaccine adjuvants to enhance the antibody and T-cell response to influenza vaccination. In a separate project, we apply forward genetics to identify genetic differences that are associated with a strong and cross-protective immune response after influenza vaccination. Vaccination of different mouse strains has identified variable immune responses after vaccination between mouse strains suggesting that genetic impact immunity to viral antigens. A systems immunology approach, including transcriptional analysis of individual T- and B-cells from these mice, will identify genes and gene-signatures associated with strong and cross-protective immunity. These gene-networks will be targets of future vaccine adjuvants to improve vaccine responses to virus vaccines.
Reassortment and genome packaging of influenza viruses
The genome of influenza virus is segmented, providing the virus with unique challenge and opportunities. The main challenge for the virus is to incorporate eight unique gene-segments into progeny virus particles. Previous studies have identified “signals” at the ends of each segment that are required for incorporation of reporter genes, but how these signals work and what they interact with is still not known. We hypothesized that gene-segment packaging required RNA duplex formation between gene-segments and that this process would eliminate viral nucleoprotein binding. To identify the viral RNA-NP interactions, we applied CLIP-seq, a state-of-the-art technology that combines cross-linking and immunoprecipitation of vRNA-NP complexes with next generation sequencing. CLIP-seq found multiple regions in the virus genome that were free or low in NP binding. These so-called low-NP binding regions were predicted to form RNA secondary structures, which we showed, were required for viral genome packaging and virus growth in vitro and in vivo.
Ongoing studies are focused on understanding how these low-NP binding regions facilitate genome packaging. We also want to understand how low-NP binding is achieved and perform biochemical analysis to improve our understanding of the RNA binding properties of viral NP. Finally, we are performing CLIP-seq on other types and subtypes of influenza viruses to understand how low-NP binding regions in H5N1 or H7N9 influenza viruses promote or limit virus reassortment and thus emergency of pandemic influenza viruses.

Bourbon virus
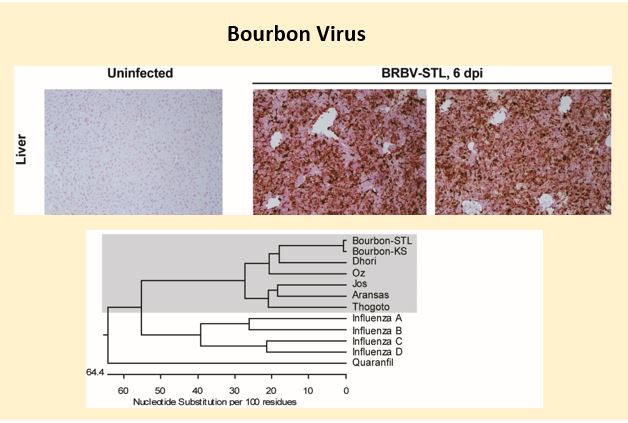
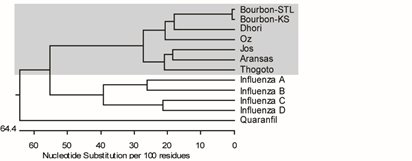
Bourbon virus is an emerging tick-borne Orthomyxovirus that is distantly related to influenza viruses. Several human cases of Bourbon virus infection have been described including two fatal cases in the Midwest region of the United States. Bourbon virus has been identified in the Lone star tick (Amblyomma americanum), which is a putative vector for transmission. The Lone star tick is endemic to the majority of the USA; raising the possibility, that significant fraction of the country has potential to be exposed to BRBV. My laboratory has cultured the second human isolate of Bourbon virus and has developed a mouse model for this novel pathogen. We are now using this model to test specific antiviral drug for efficacy against this deadly pathogen. Future studies will generate a molecular clone of this virus to (i) study the effects of genetic variation between isolates of Bourbon virus, (ii) develop a reporter virus to in vivo and in vitro testing of antiviral compounds, (iii) define the seroprevalence of Bourbon virus in people and identify the natural host of Bourbon virus in nature.
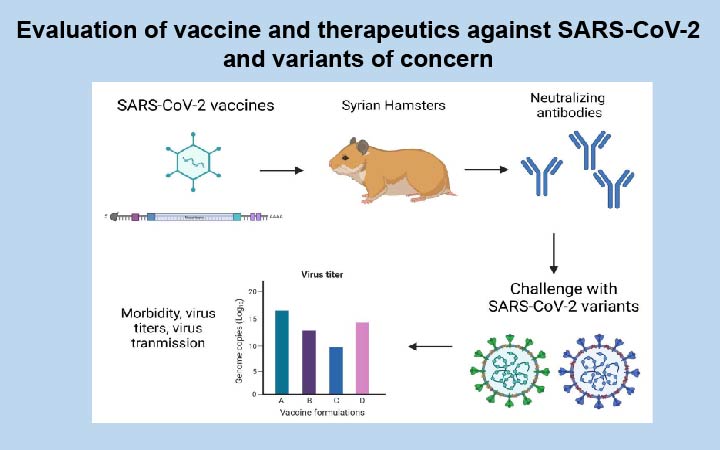
SARS-CoV-2
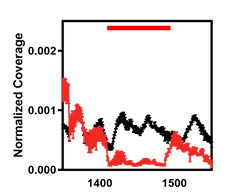
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of COVID-19 and is the first known coronavirus pandemic to mankind. As of the fall of 2020, it has infected nearly 200 million people worldwide resulting in more than 4 million deaths. Vaccine and therapeutics is needed to prevent this disease. We are testing several vaccines and monoclonal antibody based therapies against SARS-CoV-2 infection and transmission in the Syrian hamster model. We are also testing the efficacy of vaccine and therapeutics against different variants of concern including the alpha, beta and delta variant of the virus. Our future studies will focus on SARS-CoV-2 transmission, the role of spike protein adaptations and the protective effects of exiting and future vaccines against transmission of this virus.

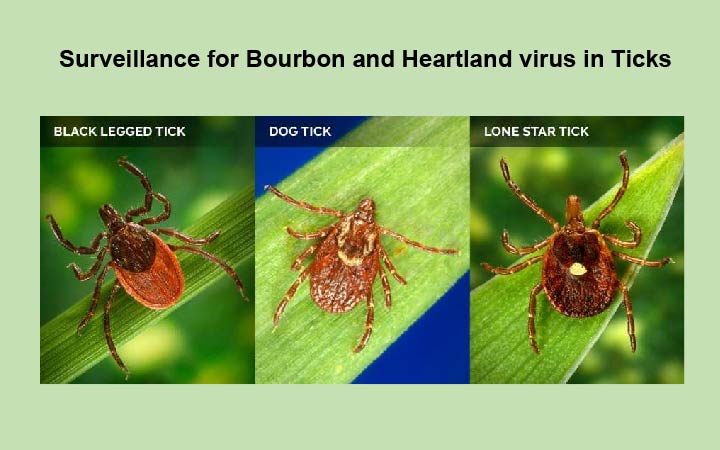
Tick Surveillance
Ticks are a well known source of many different pathogens including bacteria and viruses. Lyme disease is one of the most common tick-borne disease in the United States, however there are many other tick-borne diseases including babesiosis, ehrlichiosis, Rocky Mountain Spotted Fever, anaplasmosis, Southern Tick-Associated Rash Illness, Tick-Borne Relapsing Fever, and tularemia. The lone-star tick (Amblyomma Americanum) is also known to transmit Bourbon virus and Heartland virus. We are interested in determining the incidence of these viruses in ticks in Missouri and other parts of the Midwest. In addition, we are determining the seroprevalence of virus-specific antibodies in humans and animals exposed to these ticks.
 Jacco Boon Lab
Jacco Boon Lab